Author: Hayley Warsinske
Editors: Molly Kozminsky, Ellyn Schinke, Irene Park
We live in a world of science and technology. Biomedical research helps improve our lives everyday by providing us with vital information about everything from hygiene to Alzheimer’s disease. Computers provide us with access to wealth of information on any subject in an instant and expedite many of our daily activities. Often these two worlds overlap and computers are also used to provide scientists with information about our own health and survival to facilitate biomedical research.
For scientists who study human health, there are two common categories that most experiments fall under: in vitro and in vivo models. The term in vitro and in vivo have Latin roots, meaning in glass and in the living, respectively. In vitro studies are performed in a laboratory using petri dishes, test tubes, and other kinds of inanimate equipment. In vivo studies are performed using living organisms such as mice, monkeys, and in some cases even human subjects.
For both of these strategies, scientific experiments require laboratory space, specialized equipment, and expensive chemicals. The overall time and cost that goes into performing these studies is tremendous. In 2016, the National Institutes of Health allocated approximately 26 billion dollars specifically to scientific research. Many biomedical scientists claim that obtaining the financial resources to perform experiments is one of the greatest hurdles to furthering discovery, as described in a series of articles by Boston University.
In addition to cost, the time and manpower required to perform experiments and analyze results can be a bottleneck for productivity. In order to reduce experimental costs, increase research efficiency, and expand the depth of our understanding, some scientists have turned to in silico studies.
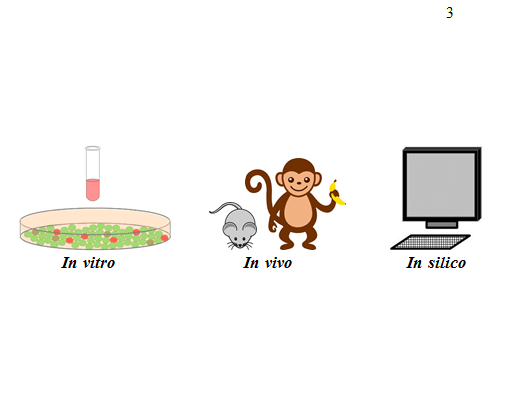
How in silico research can address the gaps of in vitro and in vivo research
In silico research offers a platform for performing experiments that would otherwise be cost-prohibitive. In silico biology, a play on in vitro and in vivo, is a strategy using math and computer science to study biological processes through computer simulations. In silico models are like the in vitro and in vivo models we discussed previously, but in place of a petri dish or mouse, experiments are performed using a computer. Experiments are performed on virtual cells, mice, and people that are simulated and analyzed by computers. Scientists can build computer programs that reflect biology and simulate biological events, and then test hypotheses by performing experiments using these programs.
Since the accessibility of computers has improved productivity and streamlined so many aspects of everyday life, it’s only logical that extending their application to biomedical studies could expedite discovery and have an overall positive impact on human health. What do these computer programs look like and how do they work? Well, like in vitro and in vivo experiments, these programs can take all shapes and sizes, modeling everything from fruit fly development to community level social behavior.
Modeling tuberculosis in lungs using computers
Since there are so many different in silico models, let’s focus on one example. Scientists at the University of Michigan have constructed an in silico model to study tuberculosis. Tuberculosis (TB) is a disease caused by the pathogenic bacterium Mycobacterium tuberculosis (Mtb). Mtb infects the lungs causing a wide range of symptoms from coughing to difficulty breathing and in severe cases can result in death of the patient.
Currently, two to three billion people (about one third of the world’s population) are infected with Mtb and about two hundred million people show symptoms of disease. People with TB are treated with antibiotics to improve their symptoms and prevent spreading the disease to others. There are currently ten FDA approved drugs for the treatment of TB and numerous drug regimens. The most common treatments last between six and nine months and include at least four different antibiotics. However, the use of antibiotics to treat TB is not always effective, and the reasons why the treatments fail are not always known. Antibiotic-resistant TB is a major global health concern and treatment regimens may be a contributing factor.
To improve these treatments and shorten the duration of the treatments, doctors and scientists use in vivo models and clinical trials to test new therapeutics or dosing regimens. In the case of TB, however, several challenges make in vivo models extremely difficult to use. Mtb grows very slowly, making it difficult to do experiments in a timely matter. In addition, the best animal model for studying TB is a monkey, which poses some ethical hurdles for researchers, not to mention the financial burden. Clinical trials for TB can be very expensive because of the long duration of treatment and bring ethical challenges of their own. Because Mtb is difficult to work with in the lab and expensive to study in humans, it is a great candidate for in silico studies.
In an effort to answer the question “What is the best treatment for TB?” scientists in the lab of Dr. Denise Kirschner are using their in silico model of TB infection to test different treatment strategies. This model, called GranSim, is like a virtual piece of lung tissue (illustrated below in Figure 2). It has a virtual lung environment (a grid) where cells and bacteria behave like they would in a real lung. The model also contains several populations of virtual cells and bacteria. The cells and the bacteria are guided by rules that define their behavior, such as when and where they can move, how they can interact with each other, and what they can do about the bacteria in the lung. The bacteria are also guided by rules that tell them under what conditions they can grow and die. The model tracks the behavior of all the cells and bacteria over time and can provide a lot of data about the state of infection, such as the number of each cell type present over time, what each cell is doing at each point in time, and what the lung tissue looks like throughout the infection.

How in silico research can help with medical research
Because the in silico model of the lung behaves very similarly to a real lung, it can be used to test how well different antibiotic treatments work. Scientists developed virtual antibiotics to treat the in silico Mtb infected lung. They tested four different treatment regimens for 280 simulated days, which counted as one experiment. In total, they performed well over two thousand experiments. One cannot even imagine doing a study of this magnitude in monkeys!
With the data generated by these experiments, researchers were able to identify risks associated with some regimens. For instance, treatment regimens that are intermittent resulted in periods of suboptimal antibiotic concentrations at the site of infection, increasing the risk of antibiotic resistance.
Experiments performed using this in silico model can provide a lot of useful and important information but only take a fraction of the time it would take to do in vivo experiments. For in silico experiments, manpower and money are not as limiting, so experiments can be performed simultaneously and repeatedly.
But like other research approaches, there are certainly limitations to in silico models. It is not possible to infer details about completely unknown mechanisms using a model because the model is built on rules –which are defined by what we already know. To be reliable, biological models also need to be validated against biological data, which can sometimes be difficult to acquire. Finally, models are only representatives of very complex systems and inherently cannot include the full complexity of the system.
Still, in silico models are becoming increasingly popular among doctors and biomedical scientists. The recent European Society of Math and Theoretical biology held talks by scientists covering biological topics including inflammation, viral infections, cancer, tissue regeneration, human development, and pharmacology. In 2012 the INSIGNEO Institute for In Silico Medicine was established as a collaboration between the University of Sheffield and Sheffield Teaching Hospitals to expand the magnitude and impact of in silico studies on medical treatments. The institute has 45 ongoing projects exploring everything from cardiovascular disease to in silico clinical trials and it is not the only institute of its kind.
In addition to the biology, in silico models used in a wide range of environmental and social challenges, everything from climate change to military strategy. These models continue to improve the pace and efficiency of research, and may even have an influence on how you are treated at your next visit to the doctor.
About the author
 Hayley is a graduate student in Microbiology and Immunology at the University of Michigan. Her research involves using math and computer modeling to understand the progression of pulmonary diseases including fibrosis and tuberculosis. Her undergraduate degree is from the Hendrix College where she studied biology. Outside of research, Hayley enjoys running, gardening, and cooking.
Hayley is a graduate student in Microbiology and Immunology at the University of Michigan. Her research involves using math and computer modeling to understand the progression of pulmonary diseases including fibrosis and tuberculosis. Her undergraduate degree is from the Hendrix College where she studied biology. Outside of research, Hayley enjoys running, gardening, and cooking.
Image Credit for Figures 1 and 2: Hayley Warsinske
