Author: Sarah Kearns
Editors: David Mertz, Zulierys Santana-Rodriguez, and Scott Barolo
Proteins do most of the work in your body: Depending on their shape, they can digest your food, fire your neurons, give color to your eyes and allow you to see colors. Proteins follow instructions encoded in your DNA to fold into their shape, but how do they “know” what shape to fold into to perform their biological functions? What happens when they fold incorrectly?
Structure Defines Function
In every biological system, including the human body, proteins promote crucial chemical reactions to keep cells alive, such as breaking down food into energy or building the scaffold that supports the structure of the cell. Proteins are chains of amino acids, which are small molecules made of carbon, oxygen, hydrogen, nitrogen, phosphorous and sometimes sulfur atoms. Each of the 20 amino acids that make up proteins has a unique side group that gives it a specific chemical property, such as hydrophobic, polar, acidic, or basic.
When a new protein is produced, RNA, a blueprint for proteins, instructs the ribosome to assemble amino acids in the proper order. As a sequence of amino acids comes off of the ribosome, it doesn’t stay in a long line, but instead folds into a compact blob. This shape is important because the protein’s function is defined by its structure.
For example, the protein amylase, which breaks down starches into simple sugars, and the protein hemoglobin, which carries oxygen in the blood, look very different (see Figure 1). Within the amylase, complex chains of sugars called starches can weave between the red and blue domains of the protein. Amino acids surrounding the starch chemically cleave bonds to yield individual sugars. Hemoglobin, on the other hand, has inorganic groups, called hemes, where oxygen can bind. This allows hemoglobin to transport oxygen throughout the body. Without their particular structures, neither amylase nor hemoglobin can function – which means no breakdown of food and no oxygen transportation.
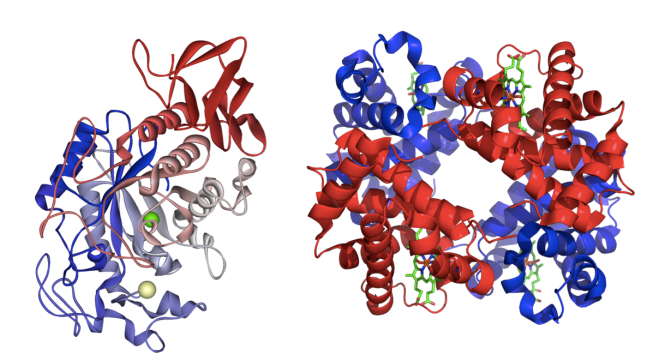
Folding into Formation
If you were to throw a string of pearls onto the ground, there is an infinite number of ways that it could land. For amino acids in a protein, this is not the case because different shapes require different amounts of energy to maintain. Chemical and physical forces push, pull, and fold each protein chain into its correct shape.
Some physical properties within a protein that contribute to this energy stabilization are hydrogen bonds, ion salt pairings, and hydrophobic interactions. Hydrogen bonds and ion salt pairings are examples of interactions between atoms. Hydrogen bonds between amino acids form the most common structural features found in proteins (see Figure 2).
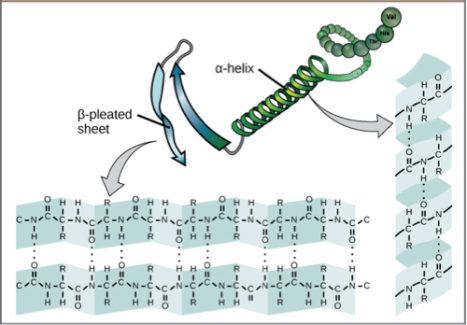
Ion salt pairings occur between charged side chains of amino acids, groups of atoms that branch off the protein backbone, sort of like how a sodium ion (Na+) pairs with a chlorine ion (Cl-) to make salt (NaCl).
But relative to hydrophobic interactions, the tendency of nonpolar molecules to be repulsed by water, the atom-atom interactions above are very weak. Hydrophobic collapse, which is caused by a protein’s interaction with water, stabilizes protein structures by pushing hydrophobic side chains toward the center of the protein (Figure 3).

Even though these forces are very important in defining structure and stability, protein structures are not static. In fact, they must be able to move quite a bit to perform their chemical functions. As a protein chain is being constructed on the ribosome, it could have a multitude of possible conformations as ion pairs can form and break. There is also a possibility that the final protein shape might change from the intermediate folds because a hydrophobic collapse may not occur until the protein chain is completed. When the chain grows, not only are there more possibilities for interactions, but also the possibilities of stronger interactions that ensure the correct structure increase. The measure of a ”correct” protein structure is described in terms of minimized energy since the structure with lower energy is more stable.
Graphs that show this minimization of energy are called energy diagrams. They plot energy levels against the possible folds, determining how much energy it takes for the protein to achieve each conformation (Figure 4).

Each time a protein folds into a more thermodynamically favored shape, the number of subsequently possible folds decreases, funneling the protein towards its lowest-energy, properly folded state.
How Protein Misfolding Can Be Dangerous
Misfolded proteins cause some diseases, like sickle cell anemia. In sickle cell anemia, a mutation in one of the amino acids in hemoglobin causes the entire protein to fold into a “sickle” instead of the ring shape of Figure 1, thus causing tissue damage and reduction in blood flow. Protein misfolds prevent important biological functions such as proper scaffolding, growth, and removal of waste. As a result, neuron growth is hampered, and toxins in the body are increased.
Typically, misfolded proteins are not in the most stable form and are instead partially-folded intermediates that are trapped in “the valleys” of the energy diagram. However, certain misfolded proteins can form very stable aggregates, called plaques, which are difficult to break up. That’s the case in some neurodegenerative diseases such as Alzheimer’s (AD), Parkinson’s (PD), and Huntington’s Diseases (HD). These misfolded aggregates – of beta-amyloid protein in AD, alpha-synuclein in PD, and huntingtin in HD – can form plaques that kill neurons in the brain.
How such accumulation leads to cell death is a mystery, but there’s evidence associating plaques with neurodegenerative diseases, diseases from brain trauma, and even diabetes. Answers to questions about protein folding and plaque formation are being investigated using computer simulations because they are too complicated for wet lab techniques.
Be sure to check out part two (coming soon) to see how games are helping to provide answers to the tricky questions about protein folding!
About the author
 Sarah Kearns is a first year in the Chemical Biology Doctoral Program at the University of Michigan. Currently, she is doing research rotations to find a long-term lab environment ideally focusing on enzyme structure-function relationships for drug development applications. Before attending UMich, Sarah got her BS in biochemistry and a minor in mathematics at the Rochester Institute of Technology where she modeled the biophysical stresses on the extracellular matrix. Her interests/hobbies other than science, are open access development, philosophy, baking bread, and hiking. You find her on Twitter (@annotated_sci), LinkedIn, or at her website Annotated Science.
Sarah Kearns is a first year in the Chemical Biology Doctoral Program at the University of Michigan. Currently, she is doing research rotations to find a long-term lab environment ideally focusing on enzyme structure-function relationships for drug development applications. Before attending UMich, Sarah got her BS in biochemistry and a minor in mathematics at the Rochester Institute of Technology where she modeled the biophysical stresses on the extracellular matrix. Her interests/hobbies other than science, are open access development, philosophy, baking bread, and hiking. You find her on Twitter (@annotated_sci), LinkedIn, or at her website Annotated Science.Read all posts by Sarah here.
Image Sources:
Figure 1: https://commons.wikimedia.org/wiki/File:Salivary_alpha-amylase_1SMD.png (left) and https://commons.wikimedia.org/wiki/Blood#/media/File:1GZX_Haemoglobin.png (right)
Figure 2: https://commons.wikimedia.org/wiki/File:Figure_03_04_07.jpg
Figure 3: https://commons.wikimedia.org/wiki/File:Cartoon_of_protein_hydrophobic_interaction.jpg
Figure 4: Sarah Kearns

5 thoughts on “How to Fold (and Misfold) a Protein (Part 1)”